Release date: 2017-12-20
Scientists have been trying to figure out the role of the genetic code. These DNA molecules, which are wrapped together, encode very important biological functions. Even if you don't change the DNA sequence, just give them a "makeup". For example, if you put a methyl "hat", it can make it happen. Earth-shaking changes. Studies have shown that DNA methylation is critical for a variety of biological processes, whether the gene is expressed, whether it can be stably transmitted, whether some functions can be achieved, and methylation is mixed.
If we can understand the secrets of DNA methylation, we can say that we are one step closer to revealing the source of life.
Today, "Nature Genetics" published a new study on the research group of Professor Tang Fuzhong from the School of Life Science and Technology of Peking University and Professor Qiao Jie [1]. Using single-cell sequencing technology, researchers in China have for the first time deeply analyzed the dynamic changes of demethylation and de novo methylation during early embryonic development, and the differences in parental genomic methylation, and proposed the use of methylation asymmetry. Trace the possibility of cell lineage.

From left to right, the author of this study, Tang Fuyue, Qiao Jie, Yan Liying
In this study, a total of 480 cells were measured in three categories, with an average of 8.4 GB per cell measurement and 10.8 million methylation sites, which is a huge amount of data.
As scientists have said in the article, this study paves the way for the deciphering of the secrets of DNA methylation reprogramming in early human embryos.
Focus
1 First reveals the dynamic balance of demethylation and de novo methylation during early embryonic development, and de novo methylation is concentrated in the DNA repeat region, suggesting that DNA de novo methylation inhibits transposon activity, Maintaining genomic stability plays an important role.
2 It was found for the first time that the level of methylation of the parental genome was different during the early embryonic development until the implantation stage. The methylation level of the parental genome was higher than that of the male parent, which means the potential effect of the methylation level of the maternal genome on embryo development. May be bigger.
3 For the first time, the asymmetric distribution of DNA methylation during embryonic cell division was traced back to a single cell source, suggesting the possibility of using the DNA methylation level to trace the cell lineage.
Demethylation and de novo methylation
There is no other way to figure out the secrets of methylation. Only genes can be analyzed one by one. Professor Tang Fufu and Professor Qiao Jie’s team at Peking University have been doing this kind of thing.
In 2014, two scientists joined the journal Nature, and for the first time in the world, systematically studied the DNA methylation of early embryo development [2]. Based on this research, the researchers used a genome-wide heavy sulphate (PBAT) sequencing technology that allows for the measurement of methylation levels in small amounts of DNA below 1 ng, which resolves individual embryos. The problem of non-uniformity of methylation between cells clearly reveals the DNA methylation profile at the resolution of single cells and single bases.
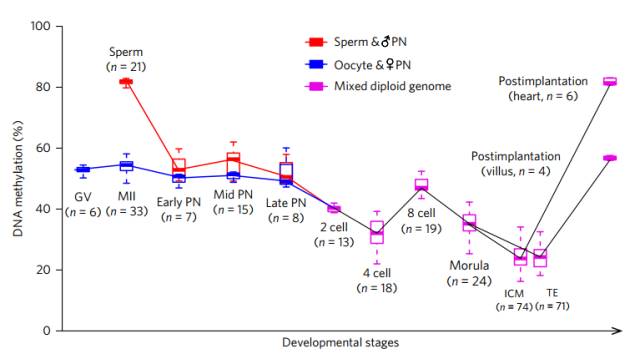
Changes in methylation levels during early embryonic development
We all know that in the early stage of embryonic development, that is, after the formation of fertilized eggs, the genome from the parents will undergo a "big cleansing" and the whole process of demethylation, which is why the genetic methylation of parents is rarely inherited. The reason for the child. The researchers found that this demethylation was carried out in three time periods.
The first large-scale demethylation occurred 10-12 hours after fertilization, when the methylation level of the paternal gene decreased from 82.0% to 52.9%, and the maternal gene decreased from 54.5% to 50.7%. . At this time, demethylation mainly occurs in the enhancer and the gene body.
The second demethylation occurred in the late fertilized egg to the two-cell stage, when the methylation level of the genome decreased from 49.9% to 40.4%; the third demethylation occurred in the eight-cell stage to the mulberry stage, the genome The level of methylation decreased from 47.0% to 35.1%. These two demethylations occur mainly in introns and various DNA repeats (essentially transposable sequences).
Careful readers may have noticed that the data on DNA methylation levels are not continuous between these three demethylations. Because, in the middle of the three demethylated "cookies", there are also two de novo methylation "sandwiches".
In the process of fusion of the prokaryotic nucleus of the germ cells, a de novo methylation occurs; during the four-cell phase to the eight-cell phase, another de novo methylation occurs. According to the data measured by the researchers, the two genes involved in the large-scale methylation were 19,861 and 53,437, respectively!
These de novo methylated genes appear to be very purposeful, and are mainly concentrated on DNA repeats (including short-spread sequences, long-spread sequences, long-end repeats, etc.), as if deliberately suppressing the activity of these repeats, Prevent activation of transposons and maintain genome stability in this way.
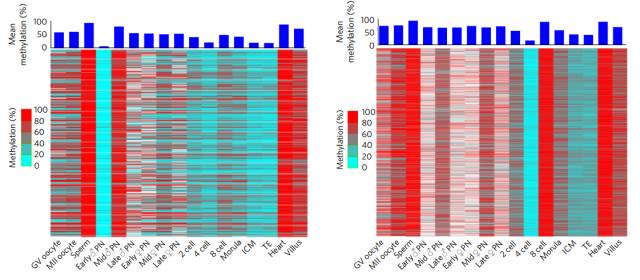
Dynamic changes of two de novo methylation genes
Interestingly, the genes that have undergone de novo methylation are not always methylated, they are likely to be restored in the next wave of demethylation, see demethylation and de novo The base is a dynamic equilibrium, and the overall methylation level exhibited by the final embryo is the product of this dynamic equilibrium equal sign.
Methylation differences between parental genes
In the process of analyzing the level of embryonic methylation, the researchers found a very interesting thing - the level of methylation between the parental genes is quite different! In the first wave of demethylation, the level of paternal methylation plummeted by 29.1%, and the female parent only fell by 3.8%!
The researchers ruled out the effects of some imprinted genes (ICRs). These genes produce differences in expression depending on the source. For example, the same allele, from the female parent, will not be expressed from the male parent. But after the exclusion, this difference still exists.
At the beginning of embryogenesis, the methylation level of the paternal gene is much higher than that of the female parent, but as the cell grows and divides, the maternal gene methylation level (23.0%) will be overtaken by the parent. This (15.2%), and this phenomenon will continue.

Dynamic changes in parental gene methylation
The researchers also measured the levels of methylation in the inner cell mass (ICM) and ectoderm (TE). The results showed that the methylation levels of the maternal and paternal in the inner cell mass were 78.6% and 76.6%, respectively, and in the ectoderm were 67.0% and 58.5%, the difference was even greater!
The researchers speculate that the asymmetry of this methylation may indicate that methylation "memory" from the female parent has a greater impact on the development of embryos, especially the placenta, and may affect the characteristics of some parent-specific genes. . However, according to the data given by the researchers, this does not seem to affect the expression of specific genes (ASEs) for the time being.
Retrospective cell lineage
Given the dynamic changes in intracellular methylation levels and the asymmetry of allele methylation, there should also be differences in methylation levels between progeny cells of different cells. If this is the case, we can track where it came from by measuring the methylation of a single cell.
To test this conjecture, the researchers made a hand to the mouse embryo. In the two-cell phase, the investigator injected one of two cells with a fluorescent label (isothiocyanate-labeled dextran, FITC-coupled dextran). Thus, only one of the two cells will illuminate, and the daughter cells that it split will also illuminate.
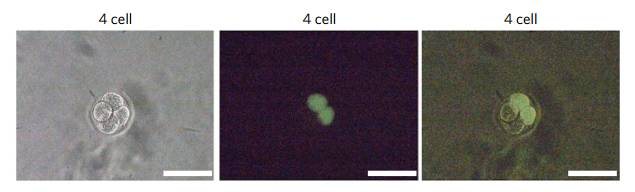
Luminescent embryonic cells, scale 100μm
When the embryos of this two-cell stage developed into the four-cell stage, the researchers sequenced the four cells separately. As a result, the cells from different sources showed a significant negative correlation. It can be seen at a glance which one is biological. . This means that using methylation to detect the origin of early embryonic cells is not a dream.
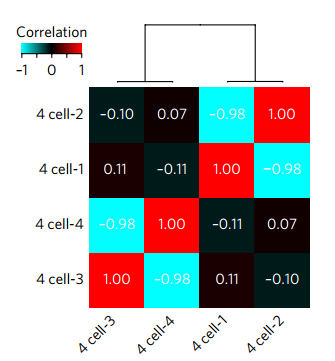
The correlation between the four cells, the more red the more "probiotic"
Dr. Zhu Ping, PhD, of the Biodynamic Optical Imaging Center of the School of Life Sciences, Peking University (now an associate researcher at the Institute of Hematology, Hematology Hospital, Chinese Academy of Medical Sciences), Dr. Guo Hongshan, Dr. Hou Yu, and a Ph.D. student at Peking University Third Hospital. The first author; Tang Fuzhong, a researcher at the School of Life Sciences, Peking University, Professor Qiao Jie from Peking University Third Hospital, and Yan Liying, a co-author of the paper. The research was funded by the National Natural Science Foundation of China, the National Major Scientific Research Program, the Beijing Municipal Science and Technology Commission, the National High Technology Research and Development Program, and the Beijing Future Gene Diagnosis Center. (Author information from Peking University Health Science Institute)
Reference materials:
[1]Zhu, P., Guo, H., Ren, Y., Hou, Y., Do, J., Li, R., Lian, Y., Fan X., Hu, B., Gap, Y ., et al. (2017). Single-cell DNA methylome sequencing of human preimplantation embryos. Nature Genetics.
[2]Guo, H., Zhu, P., Yan, L., Li, R., Hu, B., Lian, Y., Yan, J., Ren, X., Lin, S., Li, J., et al. (2014). The DNA methylation landscape of human early embryos. Nature 511, 606-610.
Source: Singularity Network
Lactobacillus Bulgaricus,Lactobacillus Bulgaricus Probiotic,Lactobacillus Bulgaricus Supplement,Lactobacillus Bulgaricus Bacteria
Biodep Biotechnology Co. ,Ltd. , https://www.biodep.com